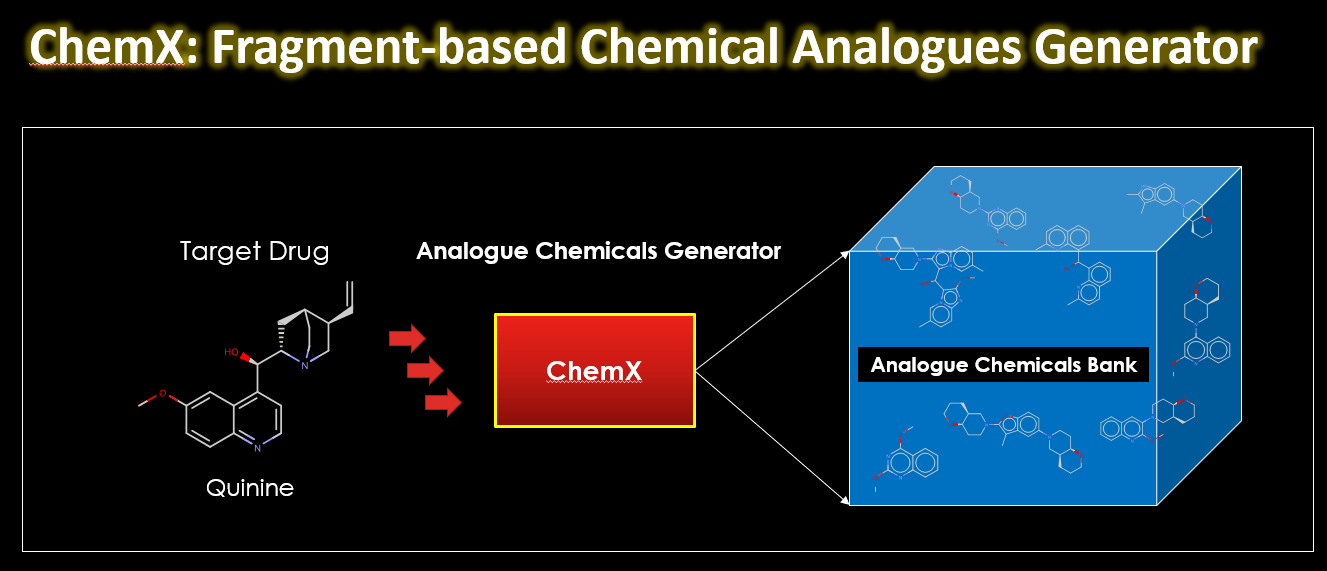
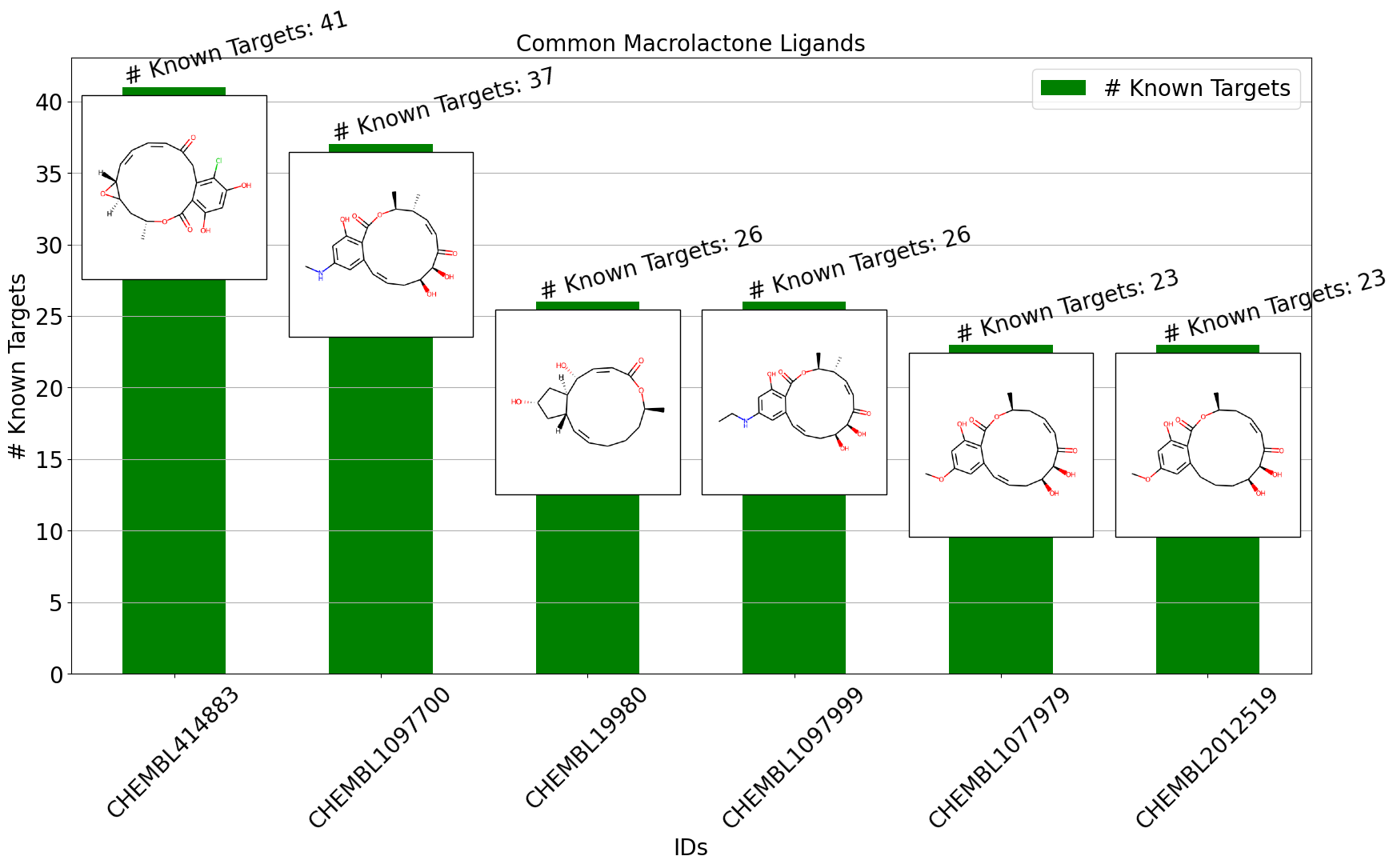
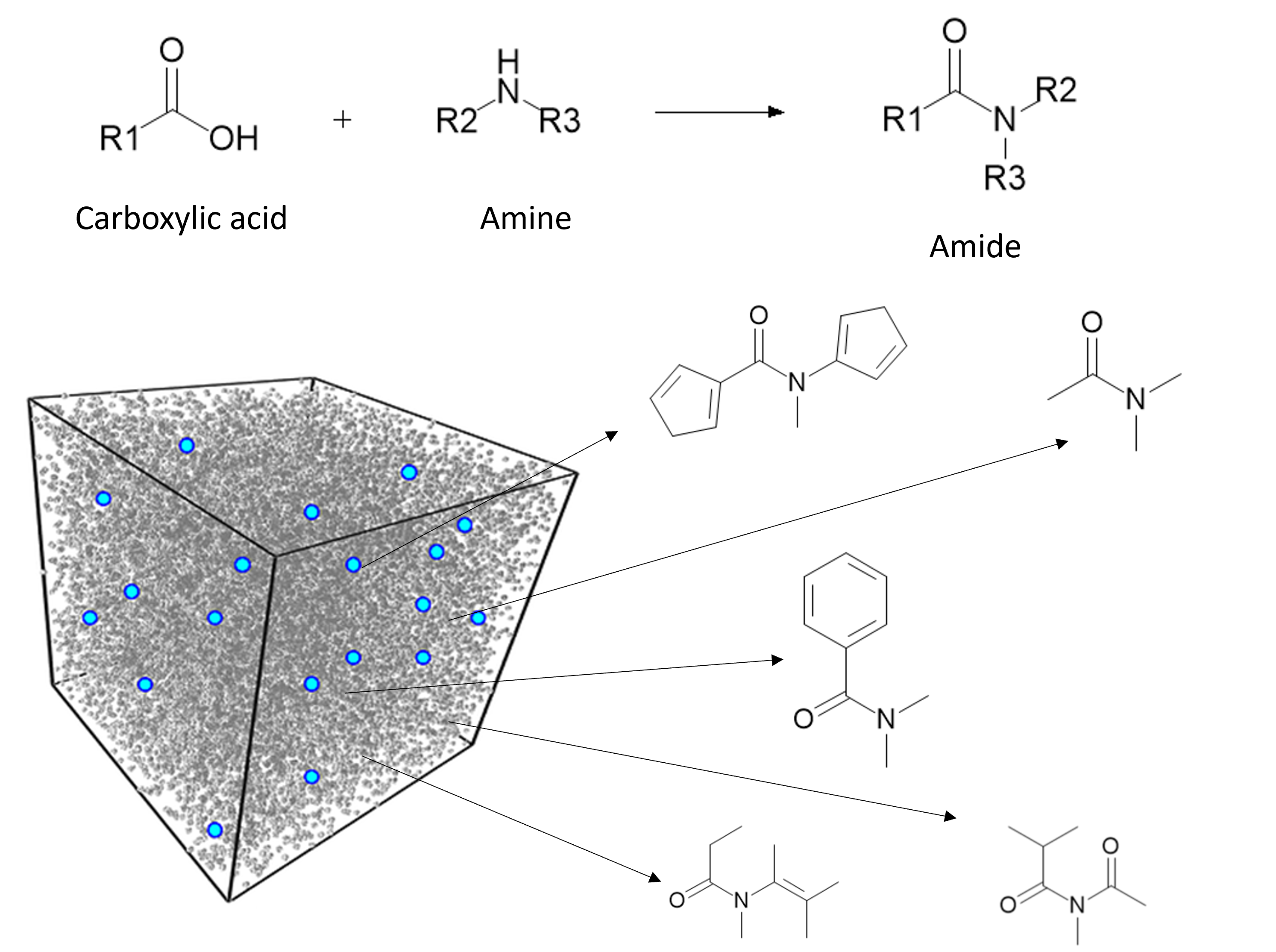
Drug discovery is a complex and resource-intensive process that involves synthesizing new compounds, often using well-established chemical reactions. While these methods have proven effective, there can be certain biases in the types of molecular structures they produce. For example, common reactions such as Suzuki or amide coupling tend to favor specific molecular frameworks, potentially limiting…