Introduction
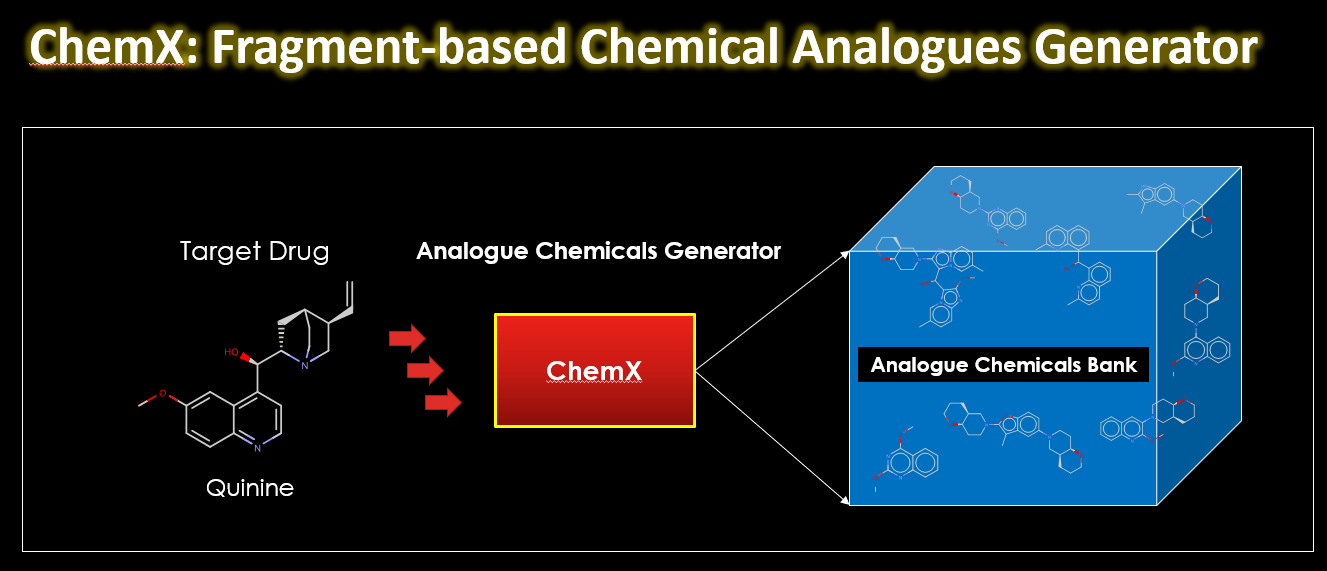
I want to introduce you to ChemX, a Python-based program I developed during a hackathon in 2019. You can use it to build virtual chemical libraries using fragment analogues to the building blocks of the target molecule. Using the RDKit library, ChemX assembles chemically similar fragments to create a virtual chemical library.
What ChemX Does
ChemX takes a target chemical compound and breaks it down into synthetically accessible building blocks. These building blocks are then replaced with others fragments with similar chemical properties. The end result is a virtual chemical library with analogs of the original compound.

How ChemX was Built
ChemX was built in Python, using the RDKit library. The initial step involved creating a chemical database containing 10,000 experimentally synthesizable molecules from the ZINC15 database. These molecules were then fragmented into synthetic building blocks to construct a ‘chemical fragments’ database. ChemX takes a SMILES string (a digital format of a chemical compound) as input, converts it into a molecular object, and performs chemical manipulations to fragment the compound. The program then searches for fragments in the ‘chemical fragments’ database with similar chemical properties, using approximately 200 2D descriptors for comparison. The chosen fragments are used to generate different arrangements, creating new virtual compounds.
What’s Next for ChemX
ChemX has a lot of room for improvement; such as implementing better approaches for merging fragments, exploring different methods for finding similar substructures, and optimizing the generated chemical library. Some of the compounds enumerated will be junk, and will need additional fine-tuning and filtering to make them drug-like. Future plans involve incorporating additional predictors, evaluating toxicity, and assessing synthetic feasibilities. The agenda also includes developing steps for compound curation, duplicate removal, and refining the controlled joining of fragments.
The Code: ChemX in Action
Note: The provided code represents the state of the ChemX project as it was during a hackathon event five years ago. While I updated the paths, I didn’t refine or update the code. However, I got it to run successfully. Use it as a starting point and consider refining it based on your specific needs.
Now, let’s dive into the code to see ChemX in action. You can also find the code related to this tutorial in this github repository.
1. Importing Libraries:
import re
import itertools
import numpy as np
import pandas as pd
from rdkit import Chem
from itertools import chain
from rdkit.Chem import FragmentCatalog
from rdkit.Chem import Descriptors
from rdkit.ML.Descriptors import MoleculeDescriptors
from rdkit.Chem import BRICS
import math
These lines import various libraries necessary for chemical processing, data manipulation, and mathematical operations.
2. ChemX Class Initialization:
class ChemX:
def __init__(self, target_compound, name):
self.target = target_compound
self.calc = MoleculeDescriptors.MolecularDescriptorCalculator([x[0] for x in Descriptors._descList])
self.chembank = open('data/' + name, 'a+')
self.templates = []
# Initialize the fragment database
self.fragment_database()
The ChemX class is defined to include the functionalities of ChemX. The __init__ method initializes key attributes, including the target compound, descriptor calculator, a file for storing generated compounds, and an empty list for templates. It also calls the fragment_database method for initialization.
3. Fragment Database Initialization:
def fragment_database(self):
# Load functional groups from file
fName = 'data/FunctionalGroups.txt'
fparams = FragmentCatalog.FragCatParams(1, 6, fName)
self.fcat = FragmentCatalog.FragCatalog(fparams)
# Load smiles from the ZINC database
zinc_file = 'data/smiles_database.csv'
zinc_suppl = [i.split(',')[1] for i in open(zinc_file, 'r').read().splitlines()][1:]
zinc_ms = [Chem.MolFromSmiles(i) for i in zinc_suppl]
# Generate synthetic fragment database
pre_synthetic_frag_database = [BRICS.BRICSDecompose(i) for i in zinc_ms]
self.synthetic_frag_database = list(set(chain.from_iterable(pre_synthetic_frag_database)))
The fragment_database method initializes the fragment database by loading functional groups from a file, reading SMILES from the ZINC database, and generating a synthetic fragment database using BRICS decomposition.
4. Fragment Writer Method:
def fragment_writer(self):
# Write synthetic fragment database to file
frg_manager = open('data/ZINC_fragments_10k', 'w')
to_write = '\n'.join(self.synthetic_frag_database)
frg_manager.write(to_write)
frg_manager.close()
The fragment_writer method writes the synthetic fragment database to a file named ‘ZINC_fragments_10k’.
5. Tanimoto Similarity Calculation:
def tanimoto_similarity(self, mol1, mol2):
# Tanimoto similarity calculation
fp1 = AllChem.GetMorganFingerprint(mol1, 2)
fp2 = AllChem.GetMorganFingerprint(mol2, 2)
return DataStructs.TanimotoSimilarity(fp1, fp2)
The tanimoto_similarity method calculates the Tanimoto similarity between two molecular fingerprints.
6. Get Similar Fragments Method:
def get_similar_fragments(self, target_cpd, n=5, threshold=0.4):
# Find similar fragments in the synthetic fragment database using Tanimoto similarity
score_frg = []
target_mol = Chem.MolFromSmiles(target_cpd)
for i in self.synthetic_frag_database:
cur_frag_mol = Chem.MolFromSmiles(i)
similarity = self.tanimoto_similarity(target_mol, cur_frag_mol)
if similarity >= threshold:
score_frg.append((i, similarity))
sorted_tuples = sorted(score_frg, key=lambda x: x[-1], reverse=True)
chosen_fragments = [k[0] for k in sorted_tuples[:n]]
return chosen_fragments
The get_similar_fragments method finds similar fragments in the synthetic fragment database based on Tanimoto similarity, with a specified threshold.
7. Fragment Target Method:
def fragment_target(self):
# Decompose the target compound into fragments
self.target_fragments = list(BRICS.BRICSDecompose(Chem.MolFromSmiles(self.target)))
The fragment_target method decomposes the target compound into fragments using BRICS decomposition.
8. Gather Fragments for All Target Fragments Method:
def gather_fragments_4alltargetfragments(self):
# Gather replaceable fragments for all target fragments
self.all_replaceable_fragments = []
counter = 0
for i in self.target_fragments:
print('Searching the database for replaceable chemical fragment ...' + str(counter + 1))
target_ds = Chem.MolToSmiles(Chem.MolFromSmiles(i))
self.all_replaceable_fragments.append(self.get_similar_fragments(target_ds, 5))
counter += 1
print('Populating all the possible fragments for different parts of your target drug ...')
The gather_fragments_4alltargetfragments method collects replaceable fragments for all target fragments.
9. Write Fragments File for Target Method:
def write_fragments_file_4Target(self, filename):
# Write replaceable fragments to file
print('Writing to file all the possible fragments for replacement ...')
header = ['fragment ' + str(u + 1) for u in range(len(self.target_fragments))]
top_row = dict(zip(header, self.target_fragments))
max_len = max(len(fragment_list) for fragment_list in self.all_replaceable_fragments)
# Create a DataFrame with a sufficient number of columns
df_columns = header + [f'extra_{i+1}' for i in range(max_len - len(header))]
cv_frame = pd.DataFrame(columns=df_columns)
# Populate the DataFrame with available data
for idx, fragment_list in enumerate(self.all_replaceable_fragments):
fragment_dict = dict(zip(header, fragment_list))
fragment_dict.update({f'extra_{i+1}': '' for i in range(len(fragment_list), max_len)})
cv_frame = cv_frame.append(fragment_dict, ignore_index=True)
# Combine top row and DataFrame
cv_frame = pd.concat([pd.DataFrame([top_row]), cv_frame], ignore_index=True)
# Save DataFrame to CSV
cv_frame.to_csv('data/' + filename + '_fragments', index=False)
The write_fragments_file_4Target method writes replaceable fragments to a file for further use.
10. Generate Fragment Templates Method:
def generate_frag_templates(self):
# Generate potential compound templates
self.potential_cpd_templates = list(itertools.product(*self.all_replaceable_fragments))
The generate_frag_templates method creates potential compound templates using the collected replaceable fragments.
11. Collect Mini Frags from Each Template Method:
def collect_mini_frags_from_each_template(self, current_template):
# Collect mini fragments from each template
mini_frags = []
for each in current_template:
num_joints = each.count('*')
numbers = re.findall(r'\d+', each)
possible_joints = ['[' + str(m) + '*]' for m in numbers]
for i in range(num_joints):
for j in possible_joints:
if j in each:
little_ones = each.replace(j, '').replace('()', ',').rstrip().split(',')
if little_ones not in mini_frags:
mini_frags.append(little_ones)
mini_frags_flattened = list(itertools.chain(*mini_frags))
return mini_frags_flattened
The collect_mini_frags_from_each_template method extracts mini fragments from each compound template.
12. Combine Fragments Method:
def combine_frag(self, max_compounds=10):
# Combine fragments to generate compounds, storing only the first 50 compounds for each input
self.generate_frag_templates()
print('Merging fragments together to generate compounds...')
for current_template in self.potential_cpd_templates:
fragms = [Chem.MolFromSmiles(x) for x in sorted(current_template)]
ms = BRICS.BRICSBuild(fragms)
for i, prod in enumerate(ms):
if i >= max_compounds:
break
sampler = Chem.MolToSmiles(prod, True)
print(i, sampler)
if sampler not in self.templates:
self.templates.append(sampler)
self.chembank.write(sampler + '\n')
The combine_frag method merges fragments together to generate compounds, avoiding duplicates in the process.
13. Main Function:
def main():
# Run ChemX for a sample chemical
mefloquine = 'OC(C1CCCCN1)C1=CC(=NC2=C(C=CC=C12)C(F)(F)F)'
print('Chemical accepted into the program.')
# Get the name for the chemical library
name = input('Name of your chemical library: ')
# Initialize and run ChemX
sample = ChemX(mefloquine, name)
sample.fragment_target()
sample.gather_fragments_4alltargetfragments()
sample.write_fragments_file_4Target(name)
sample.combine_frag()
if __name__ == '__main__':
main()
The main function initializes and runs ChemX for a sample chemical, collecting replaceable fragments, writing them to a file, and finally, combining fragments to generate compounds.
Conclusion
Thanks for checking out ChemX! This simple Python tool, born during a 2019 hackathon, offers a taste of chemical compound exploration and drug discovery using RDKit. Though in early stages, it serves as an entry point for those intrigued by the field.
I hope you find this blogpost useful. Maybe, it brings a bit of insight and maybe even helps out your drug discovery work.
And if you’re curious about related projects:
- PKS Enumerator: Design virtual macrolide libraries by permuting and adding building blocks with user-defined constraints. Read more.
- SIME (Synthetic Insight-based Macrolide Enumerator): Create in-silico macrolides with sugars, ensuring synthetic feasibility. Design libraries with specific motifs guided by biosynthetic insights. Specify a core structure, identify insertion points, and choose structural motifs and sugars. Read more.
For more content on data science, visualization, or cheminformatics, check out my other blog posts. Happy coding!