In this tutorial, I will show how to generate reaction-based molecules with python code tutorials.
I previously developed PKS Enumerator and SIME software tools which are used to design virtual libraries of macrocycles/macrolides. Both were based on a string- or template-based enumeration, and I will write a tutorial on how to do that in a later post.
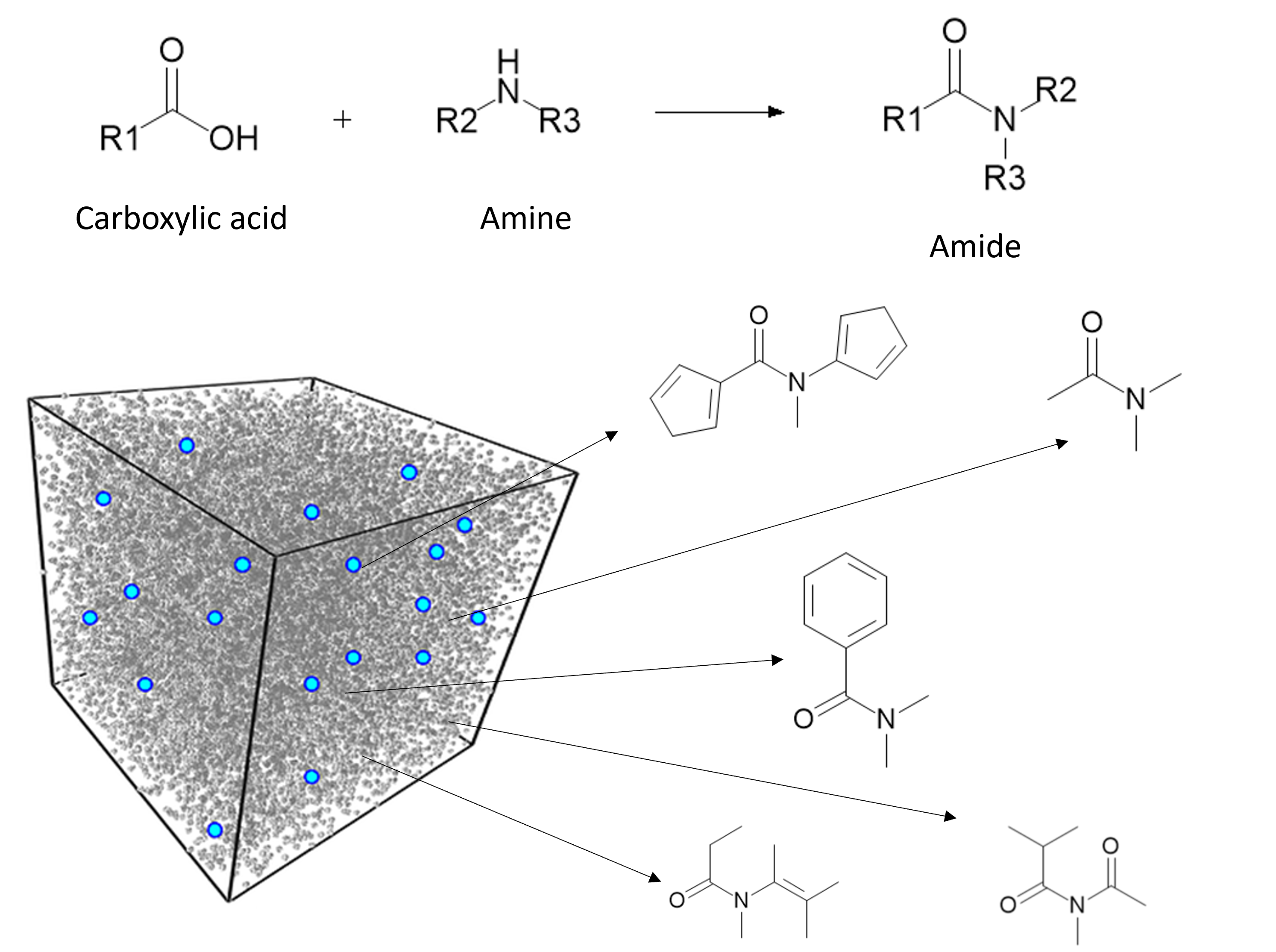
For now, I will be focusing on reaction-based transforms, such as an amide-coupling reaction. Herein, a carboxylic acid and an amine react to form an amide bond as shown in the following scheme. So the reactants are a carboxylic acid and an amine, and the product is an amide.

The focus of the blog post is on the Cheminformatics side, not synthesis or mechanism or coupling reagents or any of the lab challenges. Here, I will show how we can computationally write this reaction in python code using the RDKit library. At the end of the tutorial, we will learn how to enumerate a virtual library of possible amide molecules containing a specified N number of building blocks, given various carboxylic acids and amine reactants.
So, let’s get to the code.
If you haven’t already, install RDKit library. If you are pursuing Cheminformatics, you should definitely know this open-source library, since it is the backbone of many Cheminformatics projects and in-silico chemical manipulations. So yes, you must have that in your toolkit. I am using a jupyter notebook for this tutorial.
Let’s import the RDKit library.
from rdkit import Chem
from rdkit.Chem import Draw, AllChem
Reaction Transform (one reactant -> one product) : Ketone to Carboxylic Acid
Let’s start with a single molecule transform and see how it works. We will convert a ketone to a carboxylic acid.
Here is the reaction smart pattern and you can see the visualization of the reaction as well.
Ketone_to_CarboxylicAcid = "[Ch:1](=[O:2]) >> [C:1](=[O:2])O"
rxn = AllChem.ReactionFromSmarts(Ketone_to_CarboxylicAcid)
rxn

Reactant(s) stay on the left side of the”>>” symbol and product(s) stay on the right side. You will need to include atom numbering to explicitly specify which atom reacts in a particular way. For atom numbering, specify “: 1” ( and increment for the other atoms that you want to specify) after the element symbol and surround it in brackets ([]), such as [element symbol + modification information such as hydrogen and electric charge: number].
Now, let’s create three compounds to test this transformation.
s1 = "O=CC(CC)C"
s2 = "O=CCc1ccccc1"
s3 = "C(=O)(C(CC)C)CC"
mol_s1 = Chem.MolFromSmiles(s1)
mol_s2 = Chem.MolFromSmiles(s2)
mol_s3 = Chem.MolFromSmiles(s3)
Chem.Draw.MolsToGridImage([mol_s1, mol_s2, mol_s3])

I will refer to the compound on the left as cpd A, the middle as cpd B and the right as cpd C. We will run the transformation on them and see how the results are. Cpd A and B should run okay, but cpd C shouldn’t be converted to carboxylic acid. Below are the expected transforms for the compounds.

We will now run the ketone to carboxylic reaction on cpd A and visualize the resulting product(s). Since there is only one reactant, we just have to put the mol of cpd A (which is mol_s1) in the RunReactants function’s parameter.
resulting_mols = []
products = rxn.RunReactants((mol_s1,))
for i in range(len(products)):
current_mol = products[i][0]
resulting_mols.append(current_mol)
Chem.Draw.MolsToGridImage(resulting_mols)

The product is as expected. Now, let’s run the ketone to carboxylic acid reaction on cpd B. We just need to update mol_s1 to mol_s2 and run the jupyter notebook cell again.

This is the expected product for cpd B. Now, let’s try the same reaction for cpd C by updating mol_s2 to mol_s3. You will probably get UnidentifiedImageError because resulting_mols is empty and no product resulted from cpd 3.
Reaction Transform (two reactants -> one product) : Amide-Coupling Reaction
Now, Let’s try the amide-coupling reaction where there are two reactants and one product.
We will write the reaction smart pattern (carboxylic acid + amine => amide product) and visualize it. Note that when there are two or more chemicals on either side of the reaction, they are separated by a dot “.”
smarts = "[C:1](=[O:2])O.[Nh:3] >> [C:1](=[O:2])[Nh:3]"
amine_coupling_rxn = AllChem.ReactionFromSmarts(smarts)
amine_coupling_rxn

We will try the reaction with a simple carboxylic acid and amine. First, we will need the SMILEs and convert them to RDKit mols. We will also visualize the structures.
carboxylic_acid = "C(=O)(O)C"
amine = "NC"
carboxylic_acid_mol = Chem.MolFromSmiles(carboxylic_acid)
amine_mol = Chem.MolFromSmiles(amine)
Chem.Draw.MolsToGridImage([carboxylic_acid_mol, amine_mol])

Now, let’s run the reaction for these reactants and see if it runs okay.
products = amine_coupling_rxn.RunReactants ([carboxylic_acid_mol, amine_mol])
resulting_mol_list = []
for i in range(len(products)):
resulting_mol_list.append(products[i][0])
Chem.Draw.MolsToGridImage(resulting_mol_list)

This is the expected product, so the reaction transform went okay. Now, let’s write a function which will take in two SMILEs and return the product as SMILEs.
def amide_coupling(smile1, smile2):
mol1 = Chem.MolFromSmiles(smile1)
mol2 = Chem.MolFromSmiles(smile2)
smarts = "[C:1](=[O:2])O.[Nh:3] >> [C:1](=[O:2])[Nh:3]"
rxn3 = AllChem.ReactionFromSmarts (smarts)
products = rxn3.RunReactants ([mol1, mol2])
resulting_smile_list = []
try:
for i in range(len(products)):
resulting_smile = Chem.MolToSmiles(products[i][0])
resulting_smile_list.append(resulting_smile)
except:
pass
return resulting_smile_list
We will now try running it with a few examples. Below is the scheme with the reactants and the expected products.

Let’s call the function for Reaction A, and then visualize the product(s). We need to provide two input SMILEs for the function which will return the product SMILE(s).
cpd1 = "C(=O)(c1ccccc1)O"
cpd2 = "c1(ccccc1)N"
resulting_smiles = amide_coupling(cpd1, cpd2)
Chem.Draw.MolsToGridImage([Chem.MolFromSmiles(j) for j in resulting_smiles])

The result is the expected product for Reaction A, so it is good. Now, let’s try running it for Reaction B.
cpd1 = "C(=O)(C(C)C)O"
cpd2 = "C(=O)(NC)C"
resulting_smiles = amide_coupling(cpd1, cpd2)
Chem.Draw.MolsToGridImage([Chem.MolFromSmiles(j) for j in resulting_smiles])

This is the expected product for Reaction B, so the function seems to work okay.
With this, I will end the tutorial since we have covered some basics and a python code tutorial for reaction transforms. Hopefully, you find this post useful. Thank you so much for reading this post. As always, I welcome constructive feedback and suggestions.