Today’s tutorial is on applying Principal Component Analysis (PCA, a popular feature extraction technique) on your chemical datasets and visualizing them in 3D scatter plots.
Quick Introduction on PCA!
The following short description gives a good idea of what PCA is if you aren’t familiar with it.
Principal Component Analysis (PCA) is a linear dimensionality reduction technique that can be utilized for extracting information from a high-dimensional space by projecting it into a lower-dimensional sub-space. It tries to preserve the essential parts that have more variation of the data and remove the non-essential parts with fewer variation.
Taken from DataCamp
PCA is a linear dimension reduction / feature extraction technique commonly used in cheminformatics because there are hundreds or thousands of chemical descriptors that can characterize a molecular structure. So, we often need to use feature reduction or extraction techniques to effectively analyze and visualize see some hidden patterns among such high dimensions.
This is also often used in combination with machine learning to speed up the training, and to reduce noise and model overfitting. Here, in this tutorial, we will mainly focus on data visualization, one of the major applications of PCA.
Let’s Get to the Coding Tutorial!
The codes along with the data and the resulting figure can be located in this github repository: https://github.com/zinph/DataVisualizations/tree/master/3D_scatter_plot
First, let’s import a set of python libraries that we will need. Make sure that you already have these libraries installed before importing them.
import pandas as pd
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
from matplotlib import rcParams
from mpl_toolkits.mplot3d import Axes3D
from sklearn.decomposition import PCA
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScalerThen, we will load three csv files containing 2D RDKit molecular descriptors. These are the three datasets that we want to compare and apply PCA. Two of these datasets (ApprovedDrugs and MicrosourceSpectrum) are from the TB review project that I was helping with during my internship at Collaborations Pharmaceuticals Inc. The paper has been published here: 0.1021/acs.jmedchem.9b02075
The ‘macrolide’ dataset is extracted from MacrolactoneDB web application whose paper is also published in Nature Scientific Reports: https://rdcu.be/b4ccP.
macrolides_df = pd.read_csv('data/macrolides_2Drdkit.csv')
approved_df = pd.read_csv('data/ApprovedDrugs_20120615_2Drdkit.csv')
micro_df = pd.read_csv('data/MicrosourceSpectrum_2Drdkit.csv')Before we go further, I will show you a snippet of what one of these csv files looks like. The screenshot below is from ‘macrolides_2Drdkit.csv’ file.

Let’s check how many rows and columns are in each dataset by running the following block of codes.
print('macrolides_df : ', macrolides_df.shape)
print('approved_df :', approved_df.shape)
print('micro_df :', micro_df.shape)Below is the output from the previous code. It seems like all the datasets have the same number of columns (116) and quite different number of rows (compounds; note that each row is a compound with smiles and computed 2D RDKit molecular descriptors).
macrolides_df : (137, 116)
approved_df : (1317, 116)
micro_df : (2141, 116)Now let’s classify the datasets so that we can show each dataset separately with appropriate labels on the scatter plots. To do that, we will add a column called ‘dataset’ and fill it with :
1) ‘Macrolides’ in ‘macrolides_df’ dataframe,
2) ‘Approved Drugs’ in ‘approved_df’ dataframe,
3) ‘Microsource Spectrum’ in ‘micro_df’ dataframe.
macrolides_df['dataset'] = 'Macrolides'
approved_df['dataset'] = 'Approved Drugs'
micro_df['dataset'] = 'Microsource Spectrum'Now, all these data frames will have an extra column called “dataset” with the appropriate labels that we want to categorize: ‘TB Drugs’, ‘Approved Drugs’, ‘Microsource Spectrum’.
Next, we will prepare and process the data by imputing and normalizing. It will be easier if we merge all these separate dataframes into one. So, let’s do that.
three_merged = pd.concat([macrolides_df, approved_df, micro_df])If you run the code ‘three_merged.shape’, you will see that all the datasets are now merged and stored in the variable ‘three_merged’. It has the shape of three_merged with 3595 rows (compounds) and 117 columns (smiles, 2D rdkit descriptors and a new ‘dataset’ column that we added).
Okay, now we are about to preprocess the data. For that, we will drop the columns that we won’t need for preprocessing like ‘smiles’ and ‘dataset’.
df_descriptors = three_merged.drop(['smiles', 'dataset'], axis=1)Then, we will impute the missing values by replacing them with mean values from each column and scale each column from 0 to 1 using MinMaxScaler.
imputer = SimpleImputer(missing_values=np.nan, strategy='mean')
scaler = MinMaxScaler()
transformed_X = scaler.fit_transform(imputer.fit_transform(df_descriptors))Now, we will apply feature extraction with PCA using scikit-learn library on this prepared numpy array and project three new features that would best represent the ~100 original features. Each principal component holds a percentage of the total variation captured from the data. In the following code snippet, we will also print out the percentages of explained variables for each of the major component.
P.S. If you want to know what data type you are working with, you can call the type() built-in python method, which returns the class type of the argument (object) passed as parameter.
#PCA
pca = PCA(n_components=3)
pca_result = pca.fit_transform(transformed_X)
three_merged['pca-one'] = pca_result[:,0]
three_merged['pca-two'] = pca_result[:,1]
three_merged['pca-three'] = pca_result[:,2]
for v in pca.explained_variance_ratio_:
print('Explained variation per principal component: {}%'.format(round(v*100,2)))The outputs from running this code section is below.
Explained variation per principal component: 34.14%
Explained variation per principal component: 14.74%
Explained variation per principal component: 8.1%The total explained variables from these three major components add up to a total of 56.98%. There is a good chunk of information loss, which is one of the known cons of feature extraction / reduction techniques like PCA.
Let’s double check how many unique values are in the “dataset” column by running the code below. This is how we will color them separately in the PCA plots so we know what dataset covers which part of the chemical space.
three_merged['dataset'].unique()The output from this code is shown below. We have three categories, which is not a surprise. We classified them that way earlier.
array(['TB Drugs', 'Approved Drugs', 'Microsource Spectrum'], dtype=object)
Next, we will split the datasets again based on these three categories by executing the code section below. It will make it easier for us when we visualize and color them in scatter plots.
macrolides_df = three_merged[three_merged['dataset']== 'Macrolides']
approved_df = three_merged[three_merged['dataset']== 'Approved Drugs']
micro_df = three_merged[three_merged['dataset']== 'Microsource Spectrum']Time to Visualize the PCA Results!
Okay, we will now visualize the three major components from PCA in 3D scatter plots and distinguish the chemicals based on different categories by various colors and marker shapes. The code on the visualization part is pretty much self-explanatory (I also provided some comments here and there). We will also save the plot as ‘3D_scatterplot_PCA.png’.
If you want to modify the figure in more depth, please check out the documentation here and adjust the code based on your needs.
colors=['b', 'r', 'g'] # set three different colors to add to the PCA plot.
fig = plt.figure(1)
ax = fig.add_subplot(111, projection='3d')
# Let's set up the first dataset of actives_df.
p1 = ax.plot(macrolides_df['pca-one'],
macrolides_df['pca-two'],
macrolides_df['pca-three'],
'o', color=colors[0], # set the first color 'b' for this dataset.
alpha = 0.6, label='macrolides', # set the legend for this dataset as 'actives'
markersize=3,
markeredgecolor='black',
markeredgewidth=0.1)
# Let's set up the second dataset of micro_df.
p2 = ax.plot(micro_df['pca-one'],
micro_df['pca-two'],
micro_df['pca-three'],
'X', color=colors[1], # set the second color 'r' for this dataset.
alpha = 0.6, label='microsource spectrum', # set the legend for this dataset as 'microsource spectrum'
markersize=3,
markeredgecolor='black',
markeredgewidth=0.1)
# Let's set up the third datset of approved_df.
p3 = ax.plot(approved_df['pca-one'],
approved_df['pca-two'],
approved_df['pca-three'],
'p', color=colors[2], # set the third color 'g' for this dataset.
alpha = 0.6, label='approved drugs', # set the legend for this dataset as 'approved drugs'
markersize=3, markeredgecolor='black',
markeredgewidth=0.1)
# We will then label the three axes using the percentages explained for each major component.
ax.set_xlabel('PCA-1, ' + str(round(pca.explained_variance_ratio_[0]*100,2)) + '% Explained', fontsize=7)
ax.set_ylabel('PCA-2, ' + str(round(pca.explained_variance_ratio_[1]*100,2)) + '% Explained', fontsize=7)
ax.set_zlabel('PCA-3, ' + str(round(pca.explained_variance_ratio_[2]*100,2)) + '% Explained', fontsize=7)
fig.legend(fontsize = 'x-small', loc='upper center', markerscale=2)
plt.autoscale()
plt.rcParams["figure.dpi"] = 1000 # set the figure resolution dpi value to 1000
plt.show()
fig_name = '3D_scatterplot_PCA.png'
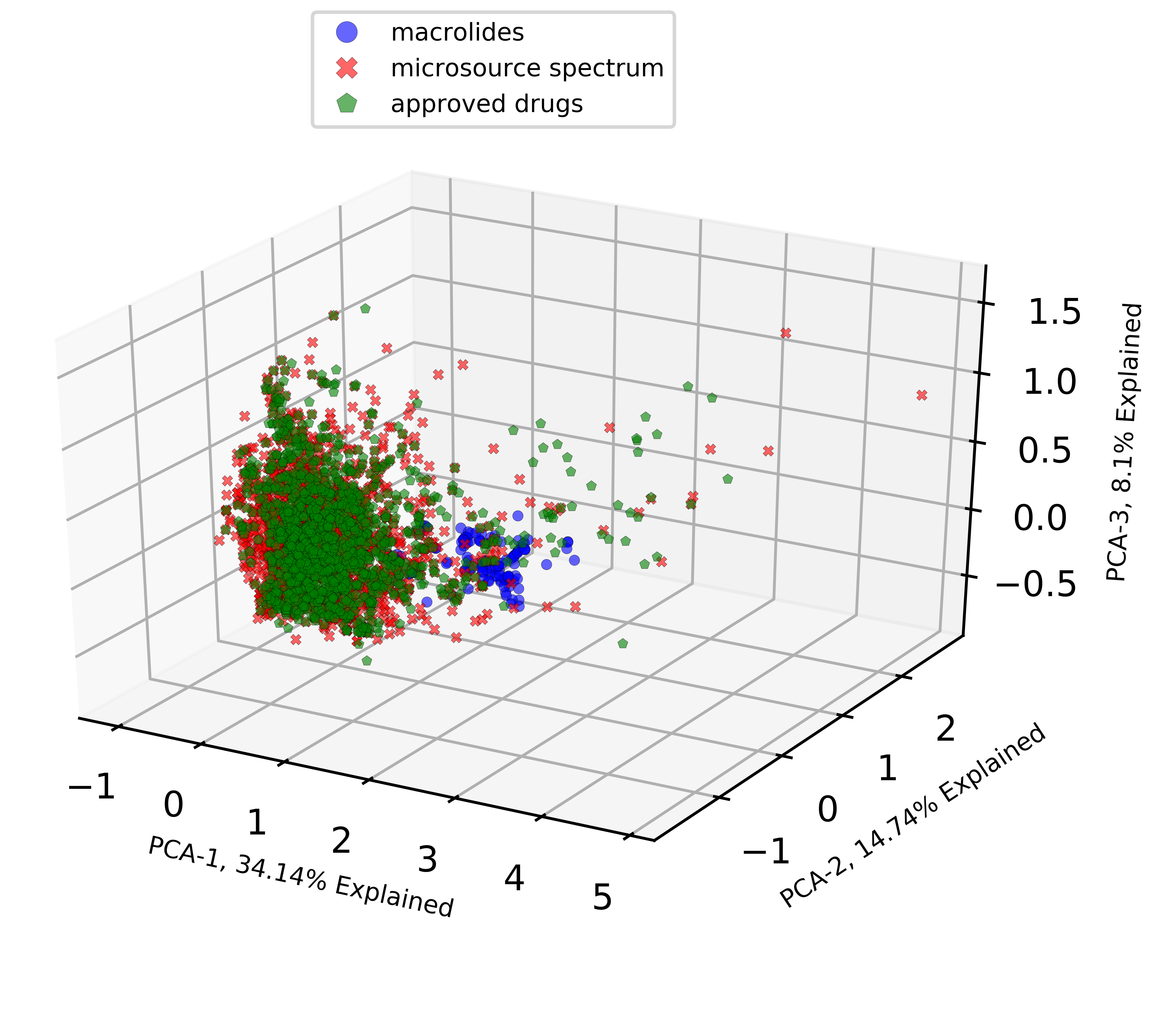
fig.savefig(fig_name)Now, you should have a resulting figure like this.

Great! We have now visualized the chemical datasets by extracting three most explainable variables with PCA and displaying them in 3D scatter plots.
So, what can we tell from a quick glance?
What this PCA analysis and visualization shows is that the ‘macrolides’ chemical dataset covers a slightly different chemical space than the other two which are ‘microsource spectrum’ and ‘approved drugs’. It makes sense because macrolides are macrolactone ring structures with slightly bigger molecular weights and unique properties than many approved drugs. Many macrolides stand apart from a majority of the two other datasets in this visualization and there are some overlapped compounds among all three datasets as well.
I won’t go much deeper into it at this point, but the overlapped compounds are quite likely macrolide therapeutics as well. But, we really have to go back and confirm that in the datasets. Of course! there is a way to single them out and visualize them but it’s for another post.
Note that there are also some ‘outlier’ molecules from ‘microsource spectrum’ and ‘approved drugs’ that may perhaps be of interest as well. These outlier chemicals do not overlap with ‘macrolides’ either so it is quite curious what type of chemical structures and properties they possess.
We also need to keep in mind that there is a certain amount of information loss when estimating their chemical coverage using PCA. We can analyze and look closer into some molecules if we have a more specific goal on this project, but for now, we have fulfilled the goal of this post which is to apply PCA on chemical datasets and and visualize them in 3D scatter plots.
I hope it has been helpful. Cheers!