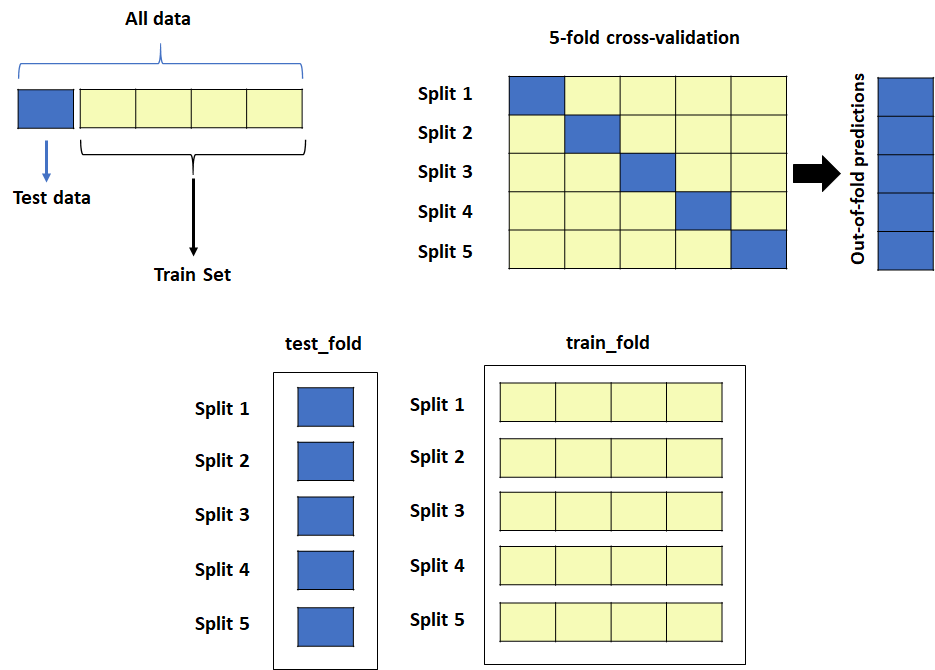
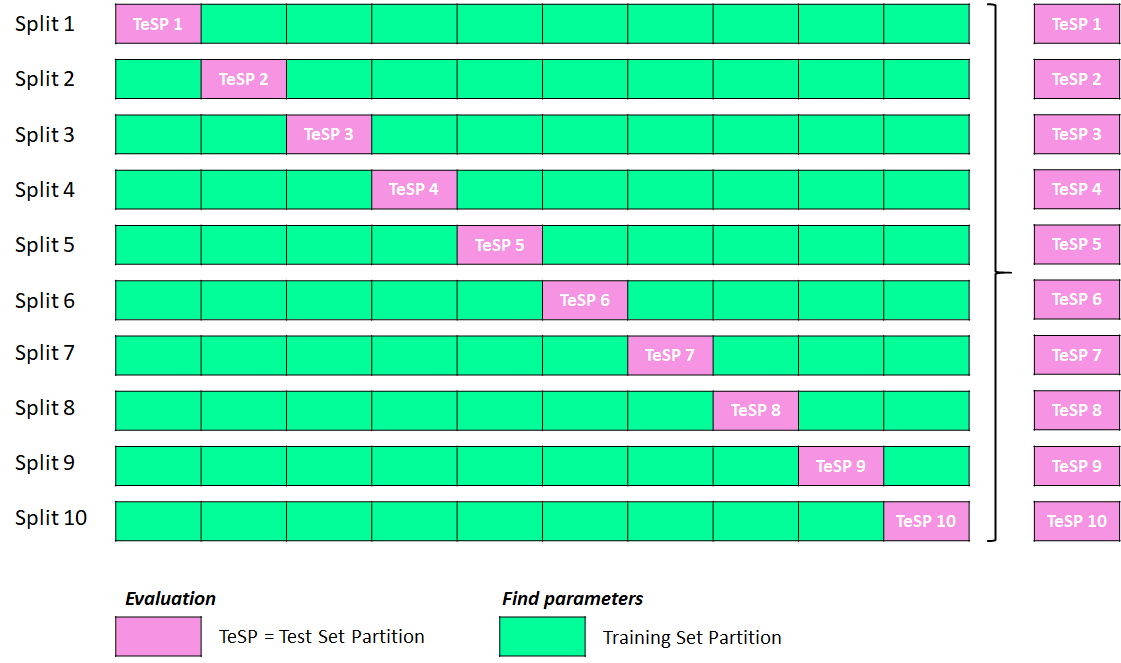
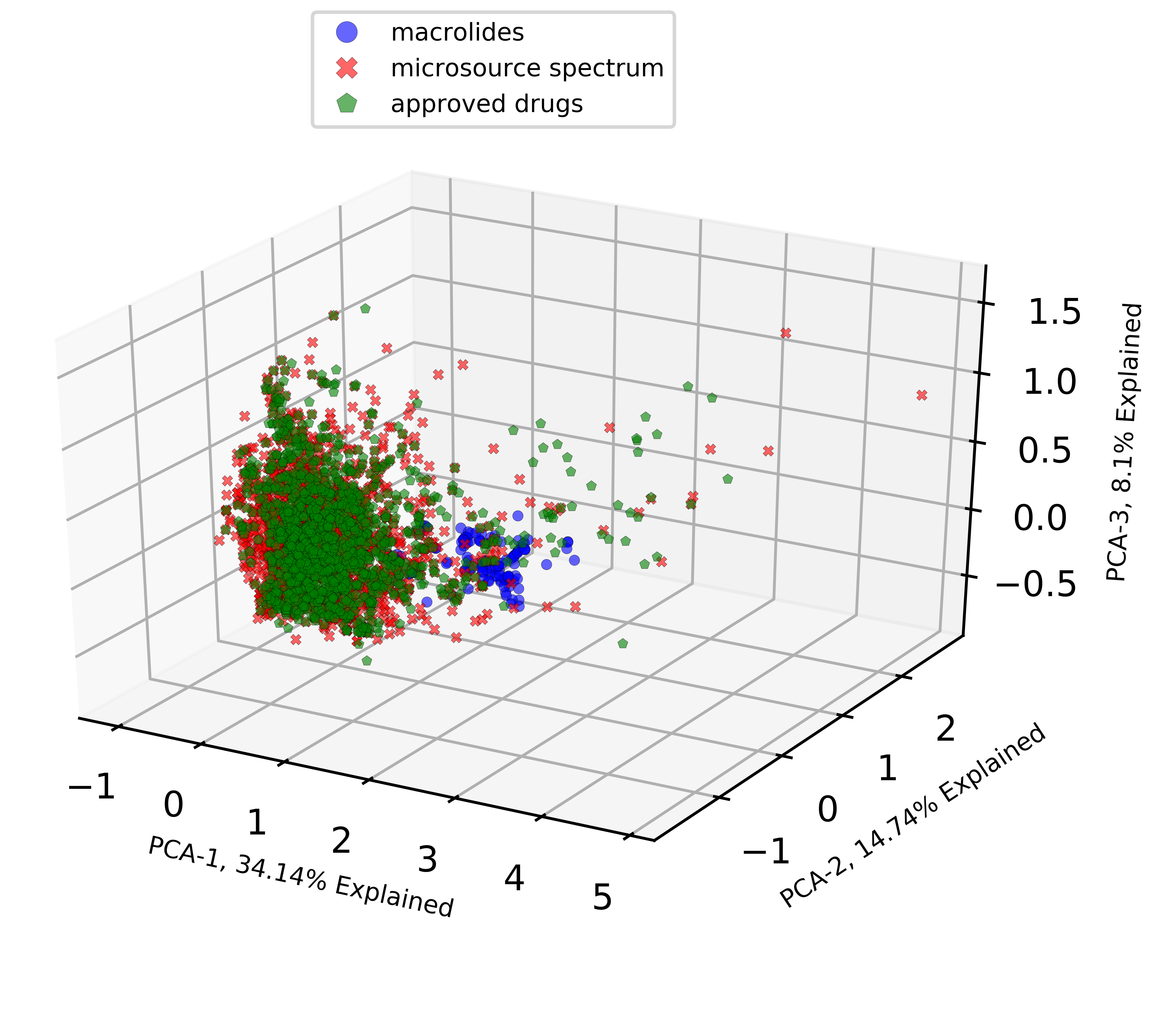
Please check out the previous blog posts from this series if you haven’t done so already: Part 1 algorithm for k-fold Cross-Validation Part 2A of the Nested Cross-Validation & Cross-Validation Series where I went through a python tutorial on implementing k-fold CV regressors using random forest (RF) from scikit-learn with a simple cheminformatics dataset with descriptors…