This is the last of the five-part series tutorial of the blog post, Computing Molecular Descriptors – Intro, in the context of drug discovery. The goal of this post to explain the python code on creating new descriptors such as MRC (developed in MacrolactoneDB study) and using Mordred descriptors. What are MRC descriptors? MRC descriptors were…
All posts tagged cheminformatics
RDKit_2D Descriptors in Python – Part 4
This is part-4 from the five-part series tutorial of the blog post, Computing Molecular Descriptors – Intro, in the context of drug discovery. The goal of this post to explain the python code on computing 2D RDKit descriptors and exporting them as CSV files. First, install the required library packages using miniconda. The code for RDKit_2D class that…
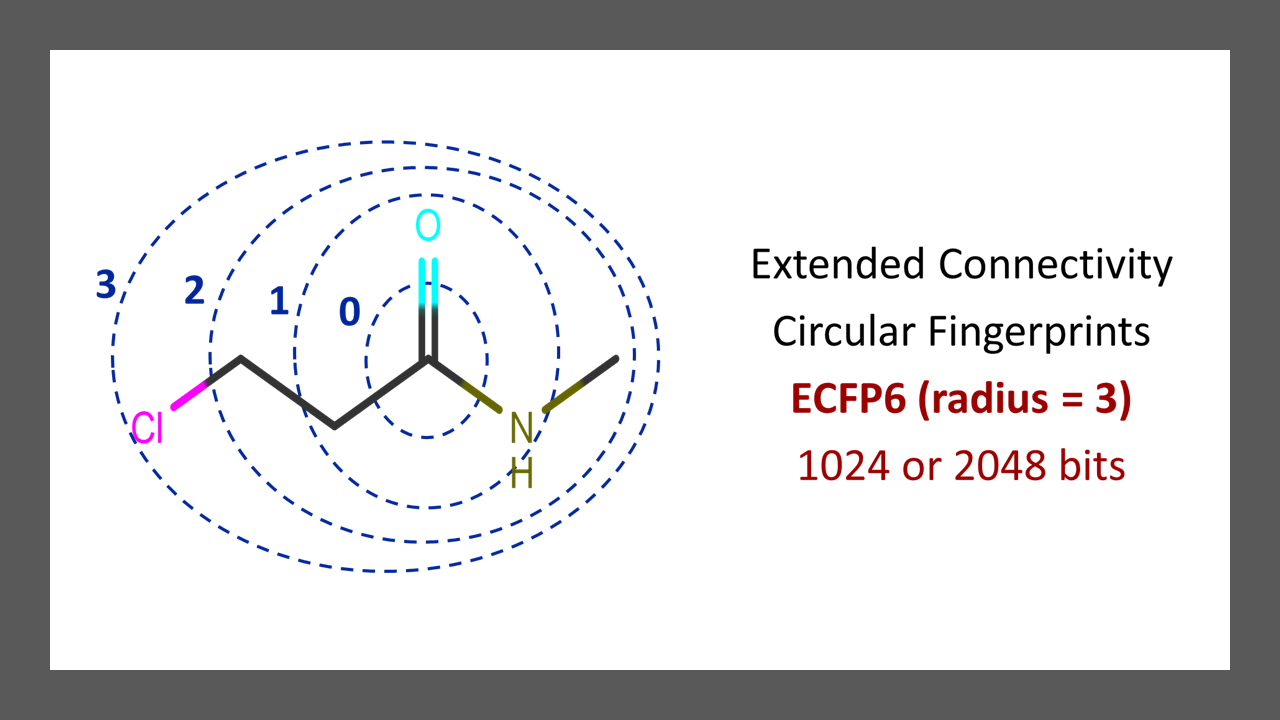
ECFP6 Fingerprints in Python – Part 3
This is part-3 from the five-part series tutorial of the blog post, Computing Molecular Descriptors – Intro, in the context of drug discovery. The goal of this post to explain the python code on computing Morgan ECFP fingerprints also known as ECFP6 (radius = 3) connectivity fingerprints. What Are ECFP Fingerprints? Please read this article and…
MACCS Fingerprints in Python – Part 2
This is from the five-part series tutorial of the previous blog post, Computing Molecular Descriptors – Intro in the context of drug discovery. The goal of this post to explain the python code on computing MACCS fingerprints. Please read this blog to familiarize yourself with MACCS. The 166 public keys (fragment definitions) of MACCS in…
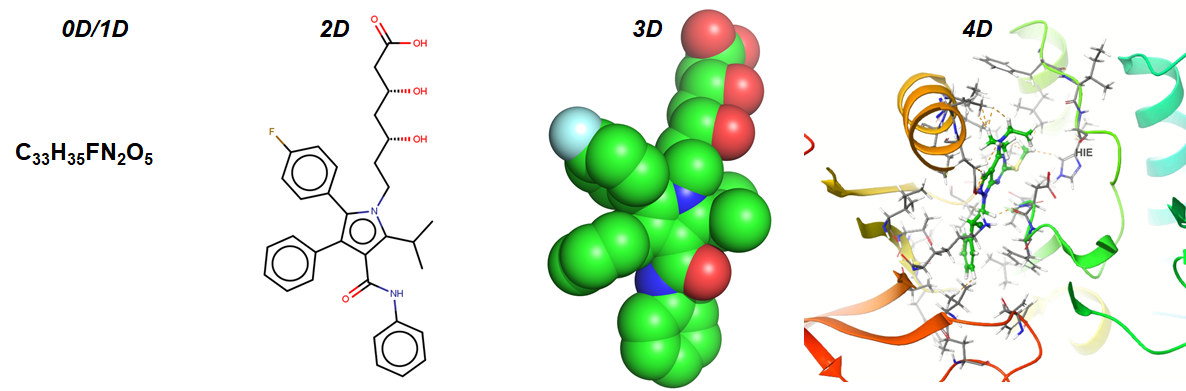
Computing Molecular Descriptors – Part 1
I will write a five-part series tutorial on implementing the python code to compute different sets of 2D molecular descriptors & fingerprints which are highly used in the context of drug discovery. Many thanks to the first-year Ph.D. students who request me to write tutorials on cheminformatics topics such as these. I welcome readers to…
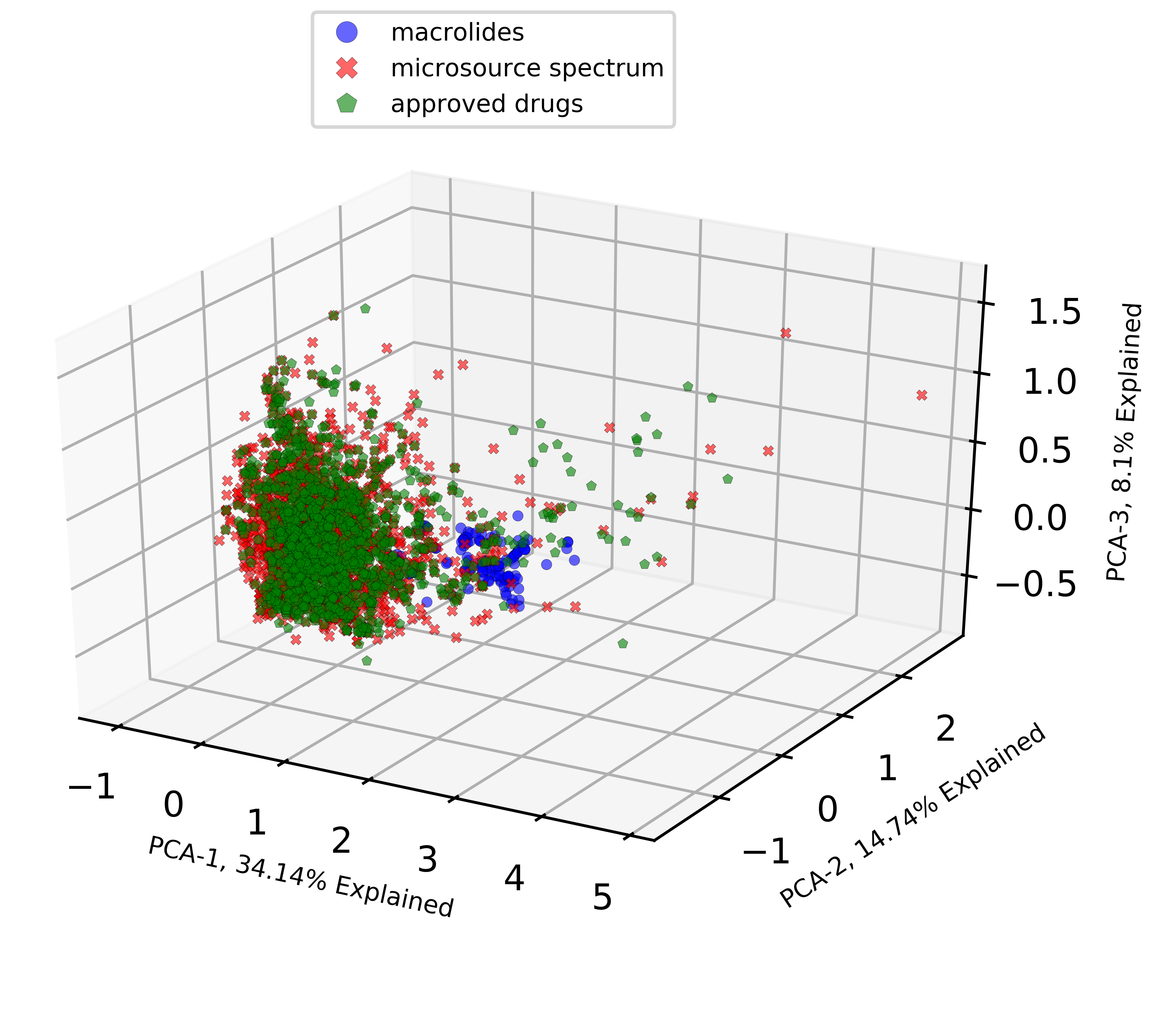
PCA Visualized with 3D Scatter Plots
Today’s tutorial is on applying Principal Component Analysis (PCA, a popular feature extraction technique) on your chemical datasets and visualizing them in 3D scatter plots. Quick Introduction on PCA! The following short description gives a good idea of what PCA is if you aren’t familiar with it. Principal Component Analysis (PCA) is a linear dimensionality reduction technique…
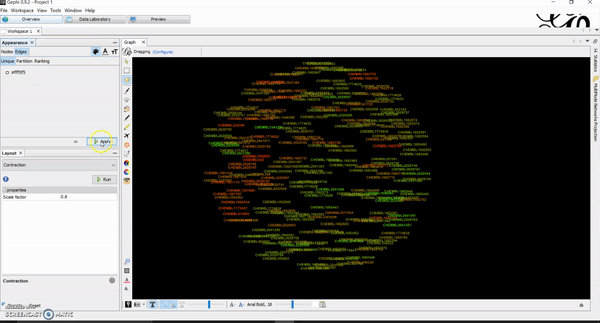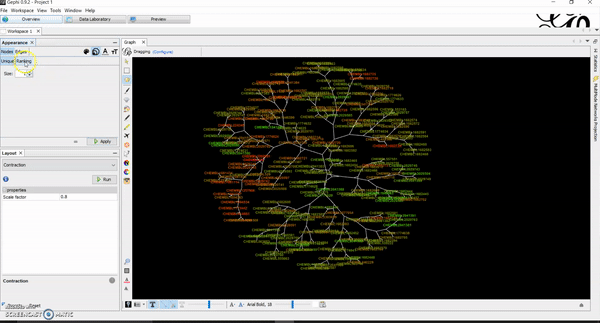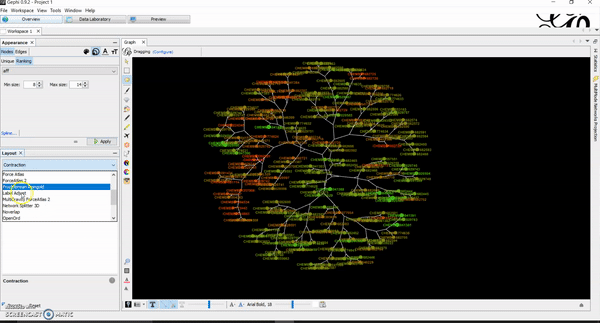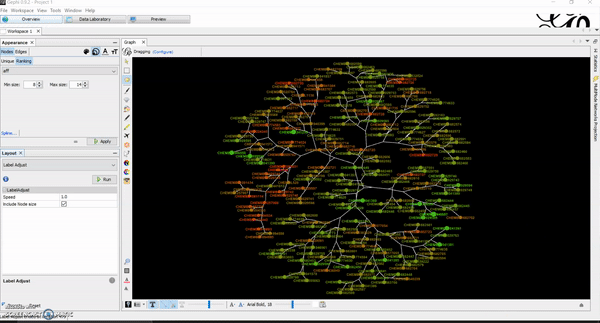
How to Generate Chemical Space Visualizations with R & Gephi
Today, I want to write a tutorial on how to generate chemical space visualizations using a combination of R and Gephi. I have found them to be a powerful way of assessing the chemical data and finding hidden patterns that could be crucial in estimating the biological endpoints of interest. Before we go on, let…
How to Compile All Mol Files into SDF file
Are you tired of creating one molecule at a time using Marvin View? I know the struggle. My folder was overflowing with individual molecule files, and it was becoming a nightmare to efficiently dock them against my target protein. But fear not, because I have found a solution, with the help of a research member…
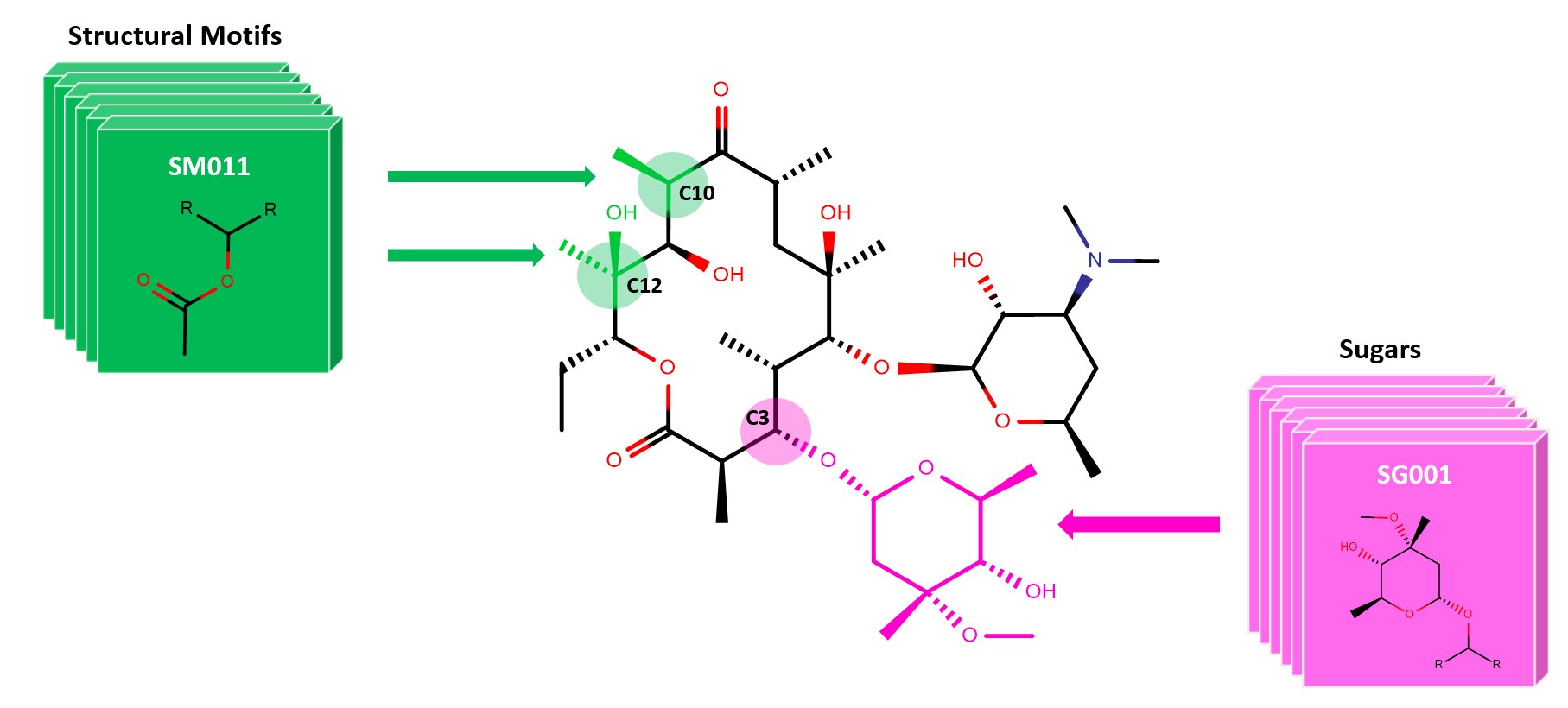
SIME: Synthetic Insight-based Macrolide Enumerator
Abstract We report on a new cheminformatics enumeration technology—SIME, synthetic insight-based macrolide enumerator—a new and improved software technology. SIME (freely available in github) can enumerate fully assembled macrolides with synthetic feasibility by utilizing the constitutional and structural knowledge extracted from biosynthetic aspects of macrolides. Taken into account by the software are key information such as…
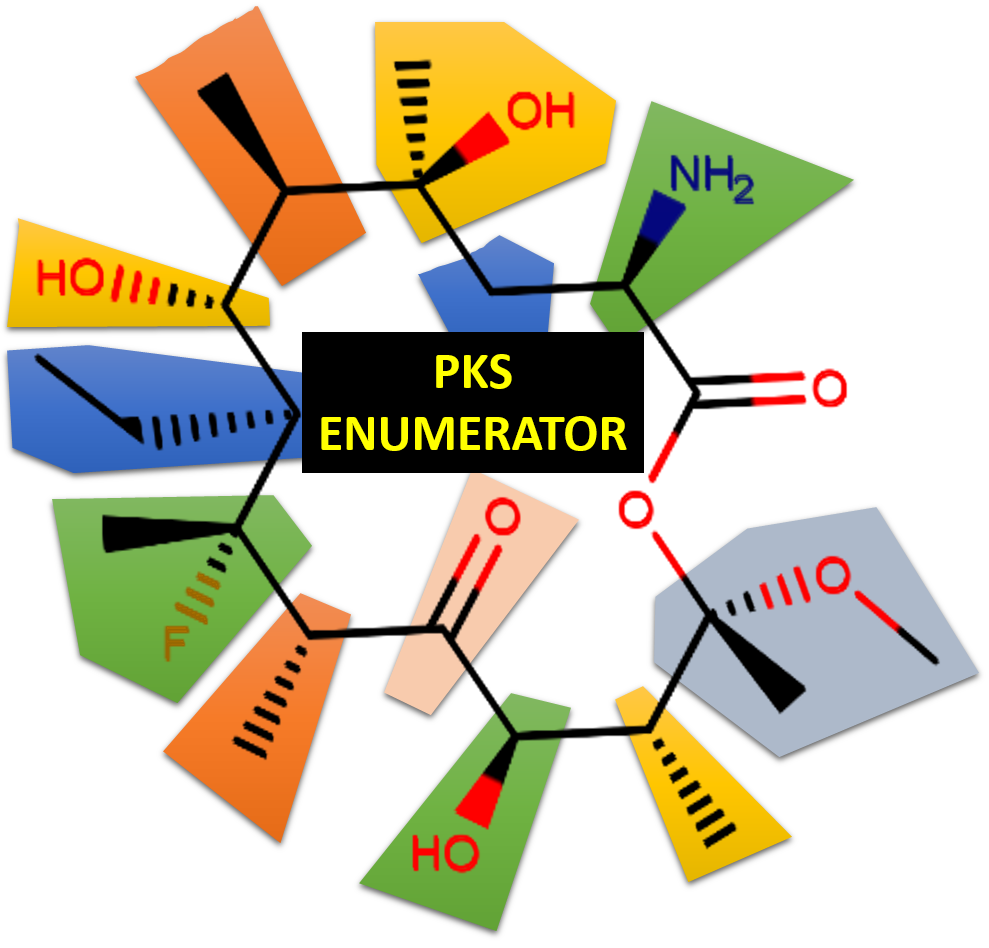
PKS Enumerator
Abstract We report on the development of a cheminformatics enumeration technology and the analysis of a resulting large dataset of virtual macrolide scaffolds. Although macrolides have been shown to have valuable biological properties, there is no ready–to–screen virtual library of diverse macrolides in the public domain. Conducting molecular modeling (especially virtual screening) of these complex molecules is…