Abstract
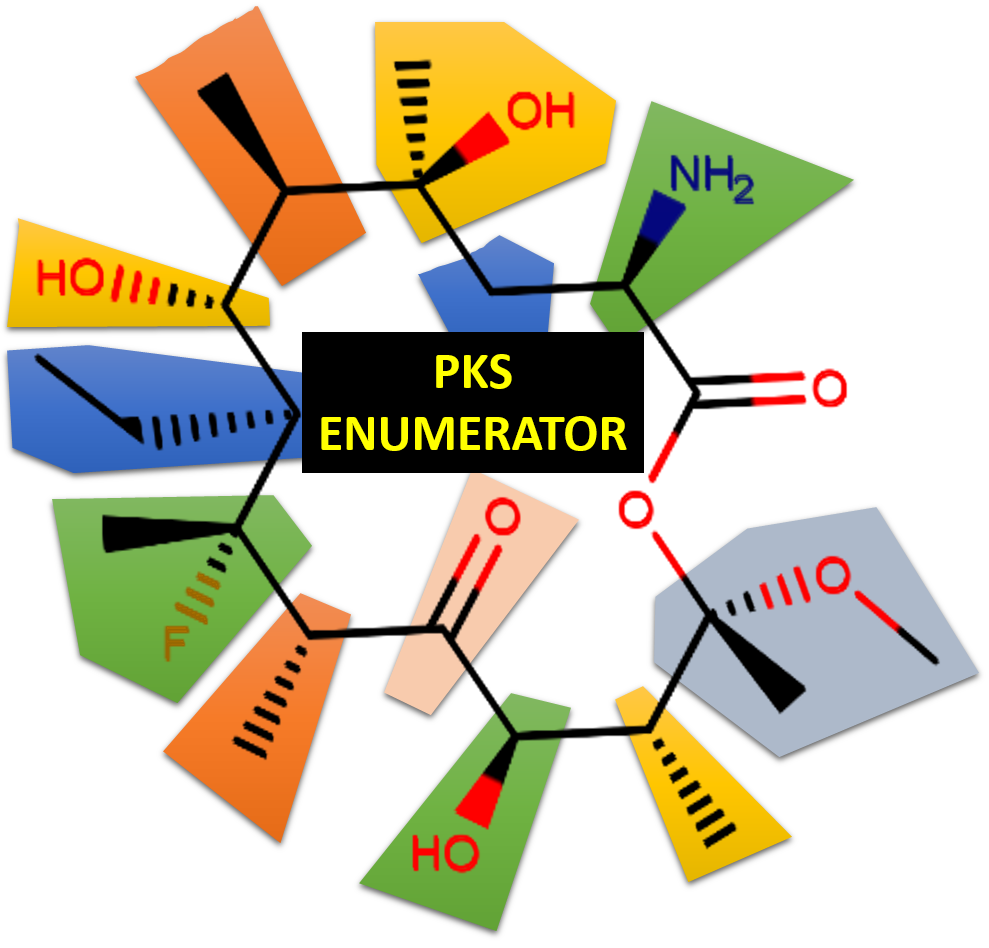
We report on the development of a cheminformatics enumeration technology and the analysis of a resulting large dataset of virtual macrolide scaffolds. Although macrolides have been shown to have valuable biological properties, there is no ready–to–screen virtual library of diverse macrolides in the public domain. Conducting molecular modeling (especially virtual screening) of these complex molecules is highly relevant as the organic synthesis of these compounds, when feasible, typically requires many synthetic steps, and thus dramatically slows the discovery of new bioactive macrolides. Herein, we introduce a cheminformatics approach and associated software (freely available in github) that allows for designing and generating libraries of virtual macrocycle/macrolide scaffolds with user-defined constitutional and structural constraints (e.g., types and numbers of structural motifs to be included in the macrocycle, ring size, maximum number of compounds generated). To study the chemical diversity of such generated molecules, we enumerated V1M (Virtual 1 million Macrolide scaffolds) library (freely available), each containing twelve common structural motifs. For each macrolide scaffold, we calculated several key properties, such as molecular weight, hydrogen bond donors/acceptors, topological polar surface area. In this study, we discuss (1) the initial concept and current features of our PKS (polyketides) Enumerator software, (2) the chemical diversity and distribution of structural motifs in V1M library, and (3) the unique opportunities for future virtual screening of such enumerated ensembles of macrolides. Importantly, V1M is provided in the Supplementary Material of this paper allowing other researchers to conduct any type of molecular modeling and virtual screening studies. Therefore, this technology for enumerating extremely large libraries of macrolide scaffolds could hold a unique potential in the field of computational chemistry and drug discovery for rational designing of new antibiotics and anti-cancer agents.
The full research article is freely available in the Journal of Cheminformatics at https://doi.org/10.1186/s13321-018-0307-6.
This project was the result of a collaboration between our computational lab of Dr. Denis FOURCHES and the biosynthetic engineering lab of Dr. Gavin Williams.
Please watch the following youtube video which explains how PKS Enumerator works.