A few people have asked me to explain and share the code for Nested Cross-Validation. I think it makes sense for me to explain the basics of whats and whys in using the NeCV first before diving into the code, so I will be covering these topics in four separate blog posts.
For part 1, I will explain the algorithm for Cross-Validation (k-fold).
For part 2, I will explain how to implement the k-fold cross-validation algorithm in python with tutorials using two cheminformatics datasets (A) a simple dataset with descriptors and endpoints of interest, and (B) high-dimensional matrices where each matrix represents features of a chemical structure (this is taken from one of my Ph.D. projects; MD-QSAR with Imatinib derivatives).
For part 3, I will explain the algorithm of NeCV and compare Cross-Validation and NeCV.
For part 4, I will write a tutorial and share the python NeCV implementation code for two datasets used in part 2 tutorial.
What is Cross-Validation?
Cross-validation is an evaluation procedure to assess the robustness and performance of machine learning models.
In traditional machine learning, you would have a training set, a validation set, and a test set. You would use the training set to fit the model and use the validation set to evaluate and tune the model hyperparameters. In other words, the validation set is used to optimize and improve the performance of the model being trained on the training set. Hence, the model has seen and learned from the validation set and you wouldn’t want to use that same validation set in the assessment of your final model.
That’s why you use the test set (also known as the external test set) for an unbiased evaluation of the final model. That test set has not been seen before by the trained model and has not been applied anywhere in the model development stage.
In cross-validation, you use the training and test set concept in a similar manner but apply it to multiple splittings rotationally to cover all your data. Hence, every entry in the original data will get to appear either in the training set or the test set. This is also known as out-of-sample testing.
Why do we often use cross-validation in Cheminformatics?
Cross-validation is often used in Cheminformatics where we do not have a whole lot of training set data, unlike in other fields such as image or voice recognition where there is an abundance of data. In a near-future where we will likely have an abundance of data for chemical/biological targets of interest, we may not likely use cross-validation or nested-cross validation much.
Having said that, applying cross-validation is an efficient use of data because you use all your data, build k different models, and then make predictions on all your data entries. In essence, you recycle your data efficiently for training, testing, and evaluating your models. It is kind of neat!
In Cheminformatics, making efficient use of limited data and assessing the generalizability of the model are the major reasons for applying cross-validation. There are several others, of course. It can be used to assess the quality and distribution in the different partitions of your data, detect some possible mistakes in the dataset, to optimize parameters (or hyperparameters) of machine learning algorithms, to reduce overfitting, and to apply other methods such as model stacking, etc.
For now, I will focus on explaining the core algorithm of k-fold cross-validation. There are also various types of cross-validation such as leave-one-out cross-validation, stratified cross-validation, and so on. But if you understand k-fold cross-validation, the others should be easy to understand.
Algorithm of k-fold cross-validation
Firstly, make sure to randomize the dataset in the beginning.
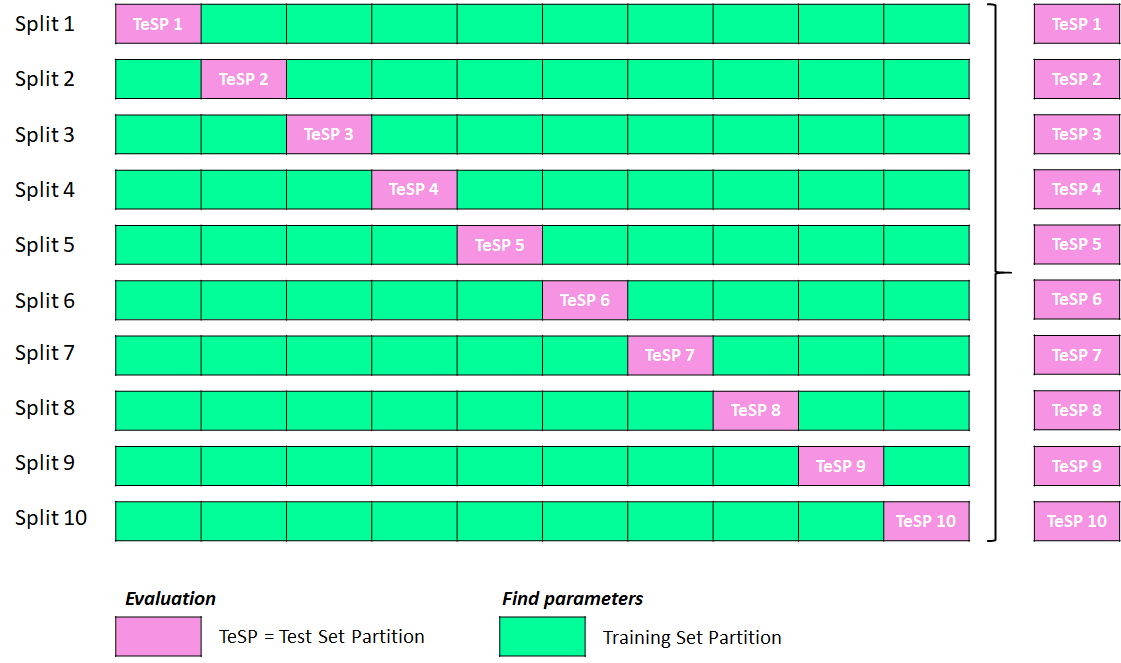
- Split the data into k partitions. In the illustration shown below, the dataset is split into 10 partitions, which makes it 10-fold cross-validation.
- Build your first model using k-1 partitions in the training set. In this specific example, since we split the data into 10 partitions, we will use 9 partitions (medium spring green color) for the training and 1 left-out partition (light pink color) in the test set. The first model you are building is for Split 1 from the illustration shown below.
- Then, move onto the next splits. Repeat the process of training other new models with k-1 training folds and making predictions on the left-out 1 test fold. This process continues until each K-fold has been predicted as the test set. In this illustration with 10-fold cross-validation, you will have created 10 different models with predictions from 10 test sets.
- Compute the average of the test set scores from those 10 test set partitions (i.e., in the illustration, it would be the predictions made on TeSP pink test blocks in each split). That will be the performance metric of your model.

I know it is a bit confusing because new models are built in each split with different training data. As you make new models in each split, people normally use the same set of model parameters that were already optimized earlier.
People normally run the cross-validation process multiple times, look at the general performance on the test set splits, adjust the parameters, and then run them again until they find a set of parameters that can produce good performance results.
In other words, the performance metric from the cross-validation models is being used to adjust the parameters, and this has been debated often due to the bias it implicitly introduces. This is because the test data would be indirectly contributing to the adjustment of the model parameters through the cross-validation process, thus more or less affecting the performance of the models. Essentially, using cross-validation to tune the models and applying it to estimate the error for the model can yield a biased estimate of the model’s performance. Thus, the nested cross-validation method was introduced to address this issue, which I will explain more in part 3 of this series.
You can read more in this article here: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-7-91
Well, that’s it for part 1 for now. Thank you so much for reading this post. I welcome constructive feedbacks and suggestions. Please do let me know if you see any mistakes or issues as well.
Don’t forget to stay tuned for part 2 where I will show tutorials in python on how to apply cross-validation with two chemical datasets.