Abstract
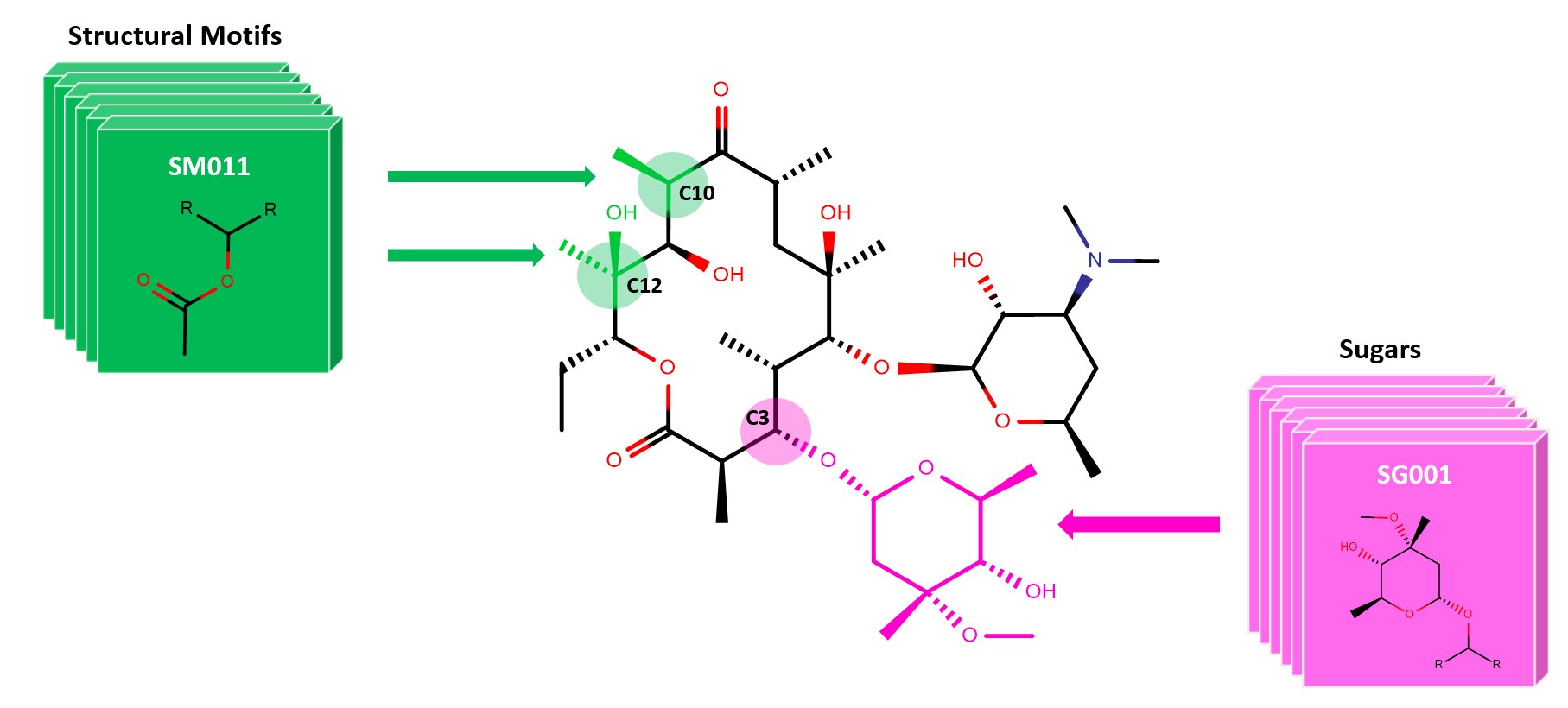
We report on a new cheminformatics enumeration technology—SIME, synthetic insight-based macrolide enumerator—a new and improved software technology. SIME (freely available in github) can enumerate fully assembled macrolides with synthetic feasibility by utilizing the constitutional and structural knowledge extracted from biosynthetic aspects of macrolides. Taken into account by the software are key information such as positions in macrolide structures at which chemical components can be inserted, and the types of structural motifs and sugars of interest that can be synthesized and incorporated at those positions. Additionally, we report on the chemical distribution analysis of the newly SIME-generated V1B (virtual 1 billion) library (freely available) of macrolides. Those compounds were built based on the core of the Erythromycin structure, 13 structural motifs and a library of sugars derived from eighteen bioactive macrolides. This new enumeration technology can be coupled with cheminformatics approaches such as QSAR modeling and molecular docking to aid in drug discovery for rational designing of next generation macrolide therapeutics with desirable pharmacokinetic properties.
The full research article is freely available in the Journal of Cheminformatics at https://doi.org/10.1186/s13321-020-00427-6 .
This project was the result of a collaboration between our computational lab of Dr. Denis FOURCHES and the biosynthetic engineering lab of Dr. Gavin Williams.
Please watch the following youtube video which describes some background on macrolides and explains how SIME works.