This is part 2A of the Nested Cross-Validation & Cross-Validation Series. I will go through a python tutorial on implementing k-fold CV regressors using random forest (RF) from scikit-learn with the first dataset: (A) a simple cheminformatics dataset with descriptors and endpoints of interest.
In Part 2B, I will cover the same python tutorial for the second dataset: (B) high-dimensional matrices where each matrix represents features of a chemical structure (this is taken from one of my Ph.D. projects; MD-QSAR with Imatinib derivatives).
Please check out part 1 of this series to learn more about the background and algorithm of k-fold cross-validation if you haven’t done so already.
Okay, so let’s dive into the code. Please note that data curating, preprocessing, molecular feature selections are usually project-based and need to be carried out. But I will not focus on them here since this tutorial is mainly for CV purposes.
I am using the following versions of the python libraries:
- python = 3.7.9
- pandas = 1.1.3
- numpy = 1.19.1
- scikit_learn = 0.23.2
Let’s import the required libraries.
import itertools
import pandas as pd
import numpy as np
from sklearn.utils import shuffle
from sklearn import preprocessing
from sklearn.impute import SimpleImputer
from sklearn.model_selection import RandomizedSearchCV
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_squared_error, r2_score
Now, we will load the dataset and shuffle the rows. Let’s also print out the pandas dataframe to see what it looks like.
The dataset along with the jupyter notebook with the code and comments can be located in this GitHub repository.
# Load the dataset and shuffle
directory = "Data/"
df = pd.read_csv(directory+"sample_dataset.csv", sep=',')
df = shuffle(df, random_state=15)
print(df)

The dataset is comprised of ID which are ChEMBL Identifiers, some 2D descriptors, and Aff (which is the log of Ki values). It has 555 entries (compounds) and 647 columns (which encode IDs, features, and endpoint).
Normally, you would do exploratory data analysis to understand your dataset better and keep the entries and molecular descriptors that would be useful in building the models. For the sake of the tutorial, we will work with the dataset as it is.
So, let’s separate out the molecular descriptors (which is everything except ID and Aff columns). Then, we will do a bit of data preprocessing and use them to build RF models.
pre_all_data = df.drop(['ID', 'Aff'], axis=1)
Then, let’s impute the data using defaults from SimpleImputer(). It will fill up missing values with the mean values of each column. Then, we will scale the data from each column to a range of zero to one using MinMaxScaler defaults.
# preprocess data, normalize columns
imputer = SimpleImputer()
imputed_data = imputer.fit_transform(pre_all_data)
scaler = preprocessing.MinMaxScaler()
all_data = scaler.fit_transform(imputed_data)
This preprocessed data is now numpy.ndarray which is ready for the application of machine learning. Please note again that this is a simplified version and you would need to put more work into the data preprocessing step.
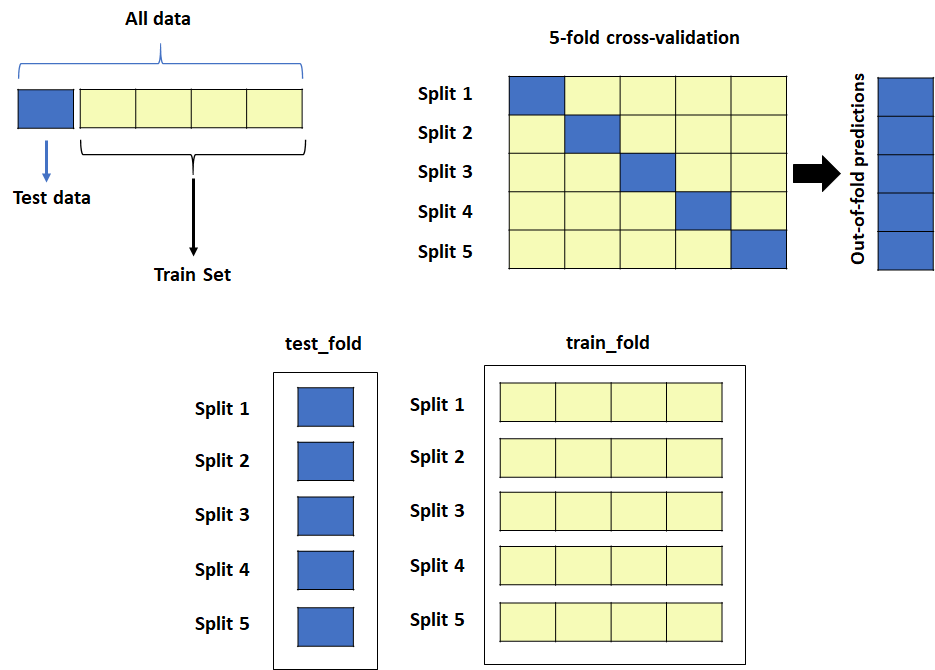
Let’s split the data into 5 partitions in this example. There are easier ways to go about it using built-in functions from some libraries, but here is the function I put together for handling the data splitting. I wrote it about three years ago, and I was recycling it from the Nested CV script and also wanted to keep track of IDs, Affs for assessing the results later.
The split_partitions function splits the data into k different folds. It requires four parameters (in the form of lists, NumPy array, or pandas series) since it is simply partitioning the data into k-folds:
a. DATA: features of the dataset. In this case, it would be molecular descriptors.
b. TARGETS: biological endpoints of the dataset. In this case, it would be Aff.
c. IDS: identifiers of the entries. In this case, it would be ChEMBL IDs.
d. folds: the number of k-fold partitions.
There will be two outputs: one folds (test sets) and k-1 folds (train sets).
They are heavily nested. The former contains a list of test sets and the later train sets. If you chose 5 fold, it will contain 5 test sets and 5 train sets. Each set is a list containing 3 lists in this order: processed data, endpoint, Ids.
I know it may be a bit confusing so I will show you an example along with a graphical illustration.
def split_partitions(DATA,TARGETS,IDS, folds):
num_val_samples = len(DATA) // folds+1
one_fold = []
nine_folds = []
for i in range(folds):
one_fold_data = DATA[i * num_val_samples: (i + 1) * num_val_samples] # prepares the validation data: data from partition # k
one_fold_targets = TARGETS[i * num_val_samples: (i + 1) * num_val_samples]
one_fold_IDs = IDS[i * num_val_samples: (i + 1) * num_val_samples]
one_fold += [[one_fold_data, one_fold_targets, one_fold_IDs]]
# prepares the training data: data from all other partitions
nine_fold_data = np.concatenate([DATA[:i * num_val_samples],DATA[(i + 1) * num_val_samples:]],axis=0)
nine_fold_targets = np.concatenate([TARGETS[:i * num_val_samples],TARGETS[(i + 1) * num_val_samples:]],axis=0)
nine_fold_IDs = np.concatenate([IDS[:i * num_val_samples],IDS[(i + 1) * num_val_samples:]],axis=0)
nine_folds += [[nine_fold_data,nine_fold_targets,nine_fold_IDs]]
return one_fold, nine_folds
Now, let’s call this function and split the data into 5 folds. The result will have test_fold (5 partitions of test set) and train_fold (5 partitions of train set).
outer_k = 5
test_fold, train_fold = split_partitions(all_data,df['Aff'],df['ID'],outer_k)
Refer to the diagram below for the graphical illustration (Figure 1). The CV algorithm was explained in part 1 of this series if you need some revision.

Essentially, the split_partitions() function is putting all the test set partitions in test_fold, and training set partitions in train_fold. Since we set outer_k = 5 (for 5-fold CV), we have five partitions in test_fold and train_fold.
Also, I wanted to retain not only processed data but also IDs and Affs. So, I made heavily nested lists. Each split in test_fold and train_fold contains 3 lists in this order: processed data, endpoints, IDs. So, in the end, the function returns something like this (Figure 2). I feel a bit uncertain about how clear it is but I hope it helps more than it confuses people.

The jupyter notebook code and files are available so you can follow the tutorial on the side and print out components of test_fold and train_fold to better understand the return of split_partitions() function.
You should also apply RF grid search for parameter tuning as you perform the cross-validation process, but for now, I will skip this step and continue with only one set of parameters. Thus, the performance from using the first random architecture is likely not gonna deliver decent results.
Note that, parameter tuning is an essential step in any model building and thus has to be included.
In the code below, we will fit each train partition with one set of RF parameters and make predictions on the relevant test partition. We iterate the process for all 5 splits and gather the results.
To keep track of endpoint targets, predictions, and IDs, I make empty lists which are outerCV_targets, outerCV_predictions, and outerCV_IDs, before starting the CV process. Then, as I go through the CV process, I append this information in smaller lists after fitting models in each split. I will just flatten smaller lists after we finish the CV process and combine the results in one single list.
outerCV_targets = []
outerCV_predictions = []
outerCV_IDs = []
for i in range(outer_k):
outer_train = train_fold[i]
outer_test = test_fold[i]
outerCV_test_data, outerCV_test_targets, outerCV_test_ids = test_fold[i][0], test_fold[i][1], test_fold[i][2]
outerCV_train_data, outerCV_train_targets, outerCV_train_ids = train_fold[i][0], train_fold[i][1], train_fold[i][2]
cv_rf = RandomForestRegressor(n_estimators= 200,
max_depth = 5,
max_features = 'auto',
min_samples_leaf = 1,
min_samples_split = 5,
bootstrap = True,
criterion="mae",
n_jobs = -1)
# Fit the random search model
cv_rf.fit(outerCV_train_data,outerCV_train_targets)
outerCV_test_predictions = cv_rf.predict(outerCV_test_data).tolist()
outerCV_predictions.append(outerCV_test_predictions)
outerCV_targets.append(outerCV_test_targets)
outerCV_IDs.append(outerCV_test_ids)
outerCV_targets_combined = list(itertools.chain.from_iterable(outerCV_targets))
outerCV_predictions_combined = list(itertools.chain.from_iterable(outerCV_predictions))
outerCV_IDs_combined = list(itertools.chain.from_iterable(outerCV_IDs))
You may want to see some performance statistics of the model so below are the root mean squared error (rms) and coefficient of determination (r2) of the cv models.
rms = mean_squared_error(outerCV_targets_combined, outerCV_predictions_combined, squared=False)
print("rms error is: " + str(rms))
r2 = r2_score(outerCV_targets_combined, outerCV_predictions_combined)
print("r2 value is: " + str(r2))
rms error is: 0.7093807455468135 r2 value is: 0.7354467885987997
You can also take a look at the scatter plot of predicted vs. experimental values here as well (Figure 3).
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.scatter(outerCV_targets_combined, outerCV_predictions_combined, edgecolors=(0,0,0))
ax.set_xlabel('Measured')
ax.set_ylabel('Predicted')
plt.show()

Now, I will put the results, experimental endpoints and IDs in a pandas dataframe and export them as csv, so you can look at individual points mapped with IDs as well.
cv_frame = pd.DataFrame()
cv_frame['IDs'] = outerCV_IDs_combined
cv_frame['ExperimentalAff'] = outerCV_targets_combined
cv_frame['PredictedAff'] = outerCV_predictions_combined
cv_frame.to_csv(directory+'RF_CV_BestModel_Predictions.csv',index=False)
People normally run the cross-validation process multiple times, look at the general performance on the test set splits, adjust the parameters, and then run them again until they find a set of parameters that can produce good performance results.
If you want to do that, you can tweak the parameters from this code within the CV loop and run that block of code in the CV loop multiple times. You can also automate this process of applying a random grid search, selecting the best parameters, and training the model. I will provide that code snippet in the NeCV python tutorial.
cv_rf = RandomForestRegressor(n_estimators= 200,
max_depth = 5,
max_features = 'auto',
min_samples_leaf = 1,
min_samples_split = 5,
bootstrap = True,
criterion="mae",
n_jobs = -1)
Doing this will mean that you are using the CV process to tune and optimize your parameters. From that series of results, you will likely choose what are the best parameters and improve your model.
Note that this process of using the CV to tune parameters and then applying it to estimate the error has been debated often due to the bias it implicitly introduces. Please also read the part 1 of this series to learn more on it.
I will stop here for now. I will post the tutorial for part 2B where I apply the same CV process with an example dataset containing high-dimensional matrices where each matrix represents features of a chemical structure. I meant to put it here but this post got longer than expected. But for part 2B, I will mainly share the code and not be that chatty because the workflow is pretty much the same.
Adios for now! As always, I thank you for visiting my blog and welcome constructive feedback. Please let me know if you detect any mistakes as well.